A.3 Arithmetic on Ranges
Sometimes you want to modify a genomic interval by altering the width of the
interval while leaving the start, end or midpoint of the coordinates unaltered.
This is achieved with the mutate verb along with anchor_* adverbs.
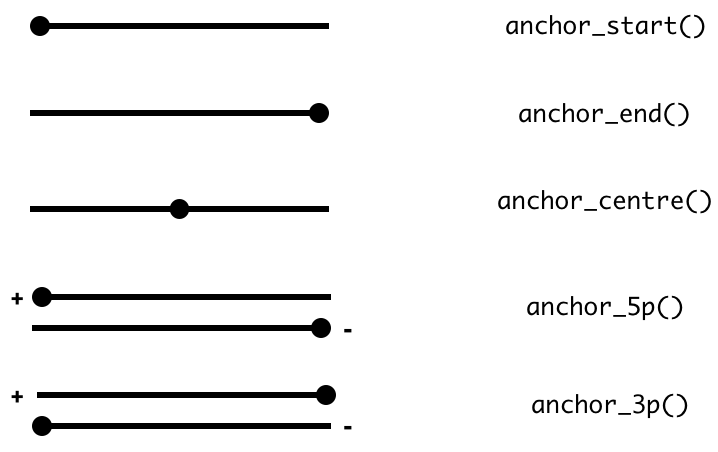
The act of anchoring fixes either the start, end, center coordinates of the
Range object, as shown in the figure and code below and anchors are used in
combination with either mutate or stretch.
#> IRanges object with 3 ranges and 0 metadata columns:
#> start end width
#> <integer> <integer> <integer>
#> [1] 1 10 10
#> [2] 2 11 10
#> [3] 3 12 10#> IRanges object with 3 ranges and 0 metadata columns:
#> start end width
#> <integer> <integer> <integer>
#> [1] 1 10 10
#> [2] 2 11 10
#> [3] 3 12 10#> IRanges object with 3 ranges and 0 metadata columns:
#> start end width
#> <integer> <integer> <integer>
#> [1] -4 5 10
#> [2] -7 2 10
#> [3] -1 8 10#> IRanges object with 3 ranges and 0 metadata columns:
#> start end width
#> <integer> <integer> <integer>
#> [1] -2 7 10
#> [2] -3 6 10
#> [3] 1 10 10#> GRanges object with 3 ranges and 0 metadata columns:
#> seqnames ranges strand
#> <Rle> <IRanges> <Rle>
#> [1] seq1 -4-5 +
#> [2] seq1 -7-2 *
#> [3] seq1 3-12 -
#> -------
#> seqinfo: 1 sequence from an unspecified genome; no seqlengths#> GRanges object with 3 ranges and 0 metadata columns:
#> seqnames ranges strand
#> <Rle> <IRanges> <Rle>
#> [1] seq1 1-10 +
#> [2] seq1 2-11 *
#> [3] seq1 -1-8 -
#> -------
#> seqinfo: 1 sequence from an unspecified genome; no seqlengthsSimilarly, you can modify the width of an interval using the stretch verb.
Without anchoring, this function will extend the interval in either direction
by an integer amount. With anchoring, either the start, end or midpoint are
preserved.
#> IRanges object with 3 ranges and 0 metadata columns:
#> start end width
#> <integer> <integer> <integer>
#> [1] -4 10 15
#> [2] -3 7 11
#> [3] -2 13 16#> IRanges object with 3 ranges and 0 metadata columns:
#> start end width
#> <integer> <integer> <integer>
#> [1] -14 10 25
#> [2] -13 7 21
#> [3] -12 13 26#> IRanges object with 3 ranges and 0 metadata columns:
#> start end width
#> <integer> <integer> <integer>
#> [1] -4 20 25
#> [2] -3 17 21
#> [3] -2 23 26#> GRanges object with 3 ranges and 0 metadata columns:
#> seqnames ranges strand
#> <Rle> <IRanges> <Rle>
#> [1] seq1 -9-5 +
#> [2] seq1 -8-2 *
#> [3] seq1 3-18 -
#> -------
#> seqinfo: 1 sequence from an unspecified genome; no seqlengths#> GRanges object with 3 ranges and 0 metadata columns:
#> seqnames ranges strand
#> <Rle> <IRanges> <Rle>
#> [1] seq1 1-15 +
#> [2] seq1 2-12 *
#> [3] seq1 -7-8 -
#> -------
#> seqinfo: 1 sequence from an unspecified genome; no seqlengthsRanges can be shifted left or right. If strand information is available we can also shift upstream or downstream.
#> IRanges object with 3 ranges and 0 metadata columns:
#> start end width
#> <integer> <integer> <integer>
#> [1] -9 -5 5
#> [2] -8 -8 1
#> [3] -7 -2 6#> IRanges object with 3 ranges and 0 metadata columns:
#> start end width
#> <integer> <integer> <integer>
#> [1] 11 15 5
#> [2] 12 12 1
#> [3] 13 18 6#> GRanges object with 3 ranges and 0 metadata columns:
#> seqnames ranges strand
#> <Rle> <IRanges> <Rle>
#> [1] seq1 -9--5 +
#> [2] seq1 -8 *
#> [3] seq1 13-18 -
#> -------
#> seqinfo: 1 sequence from an unspecified genome; no seqlengths#> GRanges object with 3 ranges and 0 metadata columns:
#> seqnames ranges strand
#> <Rle> <IRanges> <Rle>
#> [1] seq1 11-15 +
#> [2] seq1 12 *
#> [3] seq1 -7--2 -
#> -------
#> seqinfo: 1 sequence from an unspecified genome; no seqlengths