1.1 A grammar for genomic data analysis
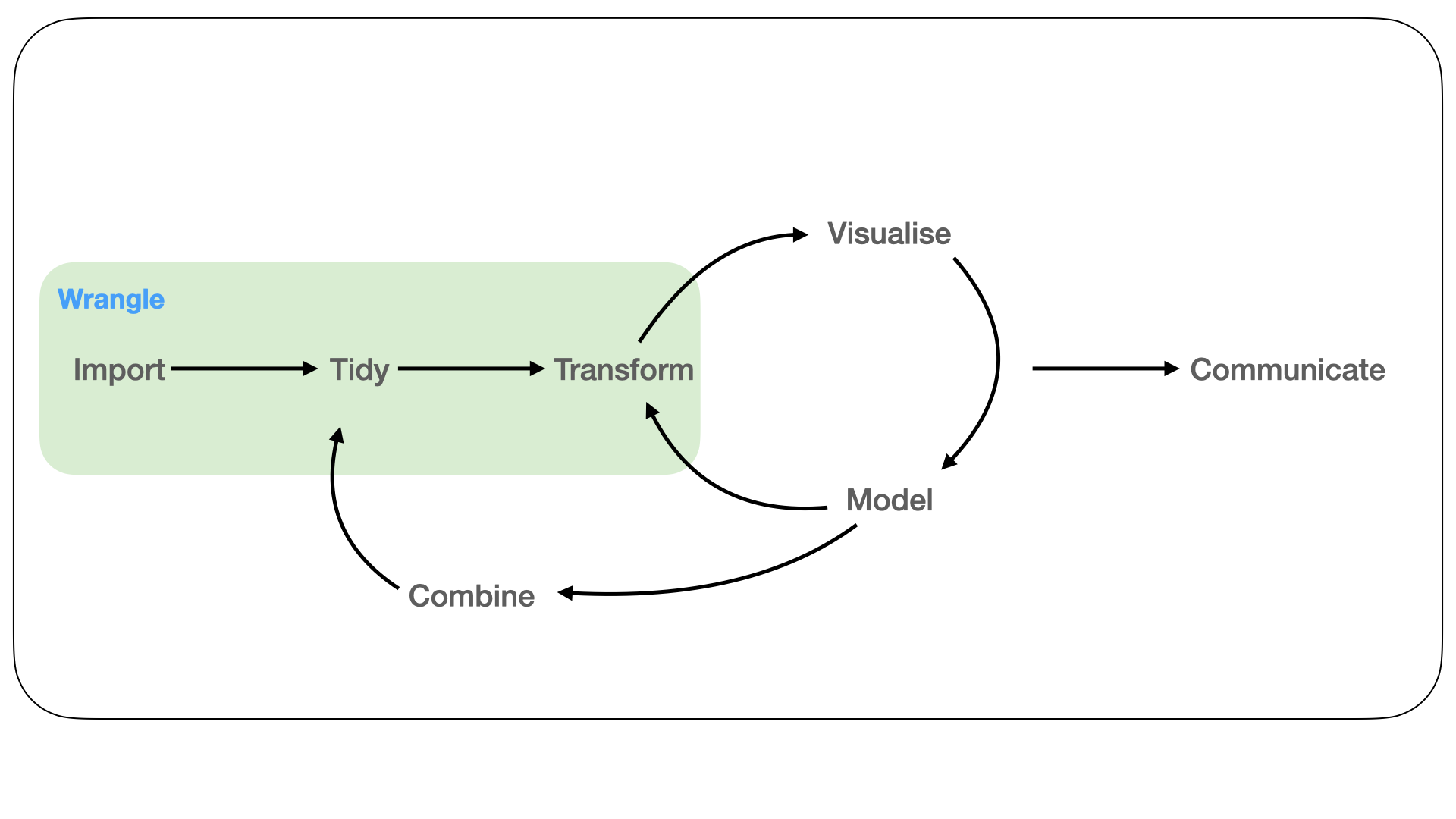
Figure 1.2: In the wrangle phase of the workflow, data from an assay is imported into a programming language. It is then tidied into a new representation that should capture the biological semantics of the measurements. Following this, the representation can be transformed to generate new summaries.
The approach taken by the suite of software packages collectively known as the tidyverse is an attempt to formalise aspects of the EDA process in the R programming language under a single semantic known as tidy data (R Core Team 2019; Wickham et al. 2019; Wickham 2014). Simply put, a tidy data set is a rectangular table where each row of the table corresponds to an observation, each column corresponds to a variable and each cell a value. There is a surprisingly large amount of utility that can be achieved with this definition. By having each column representing a variable, variables in the data can be mapped to graphical aesthetics of plots. This paradigm enables the grammar of graphics as implemented by ggplot2 (Wickham, Hadley 2016; Wilkinson 2005). User interfaces as implemented by tidyverse, and in particular the dplyr package, are fluent; they form a domain specific language (DSL) that gives users a mental model for performing and composing common data transformation tasks (Wickham et al. 2017; Fowler, n.d.).
It is unclear whether the fluent interfaces as implemented using the tidy data framework can be more generally applied and useful in fields such as high-throughput biology where domain specific semantics are required (Figure 1.2). This is particularly true in the Bioconductor ecosystem, where much thought has gone into the design of data structures that enable interoperability between different tools, biological assays and analysis goals (Huber et al. 2015a).
Chapter 2 shows that the tidy data semantic is applicable
to in memory data measured along the genome and develops a fluent interface to transforming
it called plyranges. The software provides a framework to an assist
an analyst to compose queries on genomics datasets. Our software is agnostic to how counts from bulk assays have been obtained. Indeed, we have used data obtained from both alignment and quantification based approaches throughout the thesis to perform useful analyses.
This chapter has been published as Lee, Cook, and Lawrence (2019).