1.2 Integration of genomic data structures
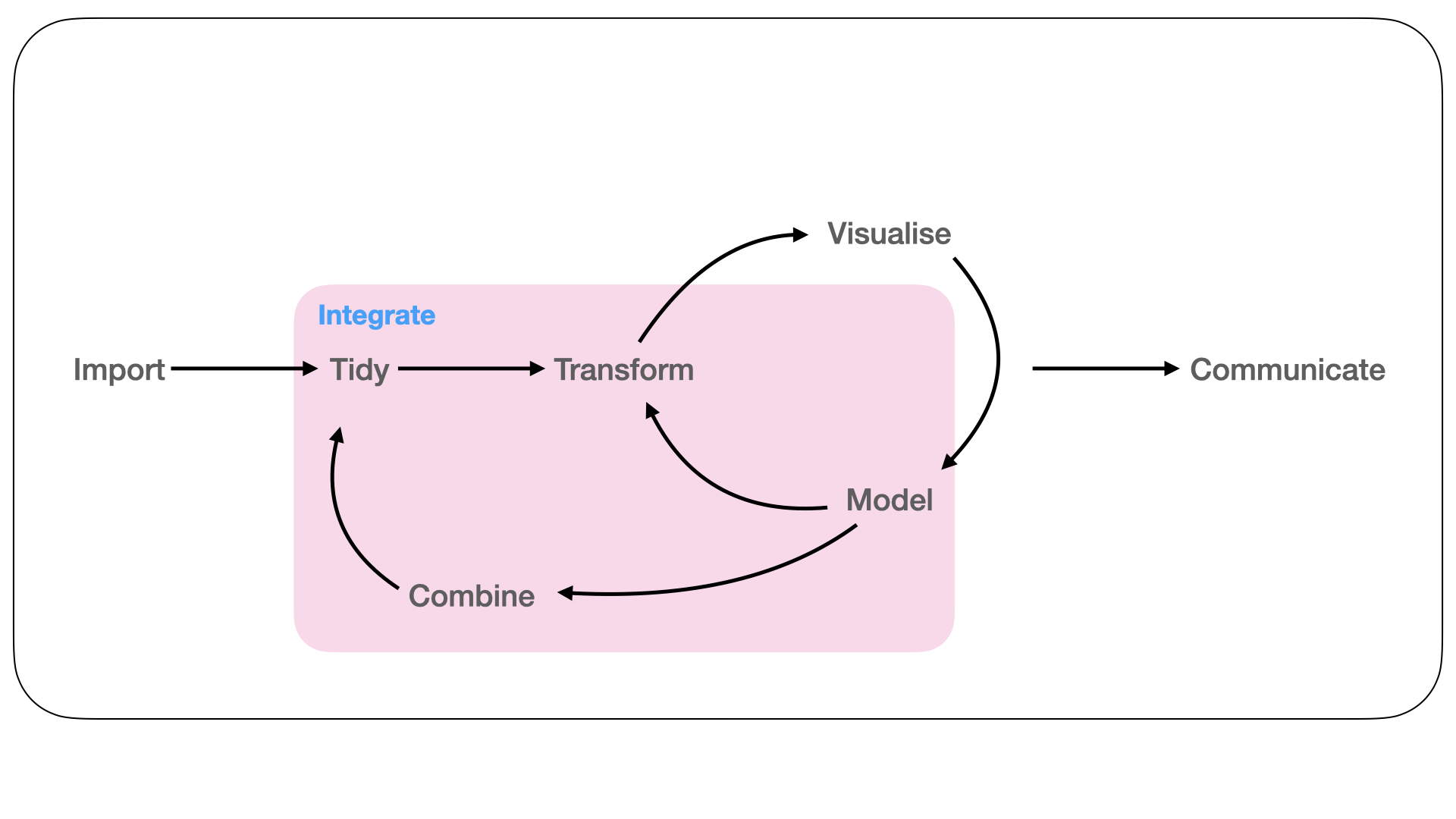
Figure 1.3: Data from multiple assays can be combined together using modelling and transformations to gain biological insight. This is achieved either via a statistical technique or more simply via joining data and model results so they have a common representation across different granularities of the genome.
It is rare that a biological data analysis will involve a single measurement assay or that only one aspect of a measurement assay will be of interest to the biological question under study (Figure 1.3). While there are many approaches to integrating data sets from multiple assays using multivariate statistical techniques (Meng et al. 2016; Stein-O’Brien et al. 2018) and data structures to represent them (Ramos et al. 2017), there has been little thought given to the interoperability between these approaches and the tidyverse. In Chapter 3 we describe a simple end-to-end workflow for integrating results along the genome using plyranges. This workflow shows that our grammar based approach does not impair interoperability between the tidyverse and Bioconductor approaches, and in fact they work seamlessly together. This chapter has been published as S. Lee, Lawrence, and Love (2020).